Designing disease-modifying strategies
To elucidate the molecular mechanisms underlying vulnerability of neurons to protein misfolding and degeneration, we are using proteomic (Hosp et al., 2017; Riera-Tur et al., 2022) and single cell RNA sequencing (Fig. 1) approaches in neurodegeneration models. These analyses enable us to identify candidate genes and pathways that can be targeted to increase neuronal resilience and ameliorate disease symptoms. To explore the therapeutic potential of the identified candidates, we then perform loss- and gain-of-function studies in cell culture and mouse models. We use transfection and lentiviral transduction of primary neurons (Fig. 2) and stereotactic viral injections into the mouse brain (Fig. 3), followed by histological, biochemical and behavioral phenotypic analyses.
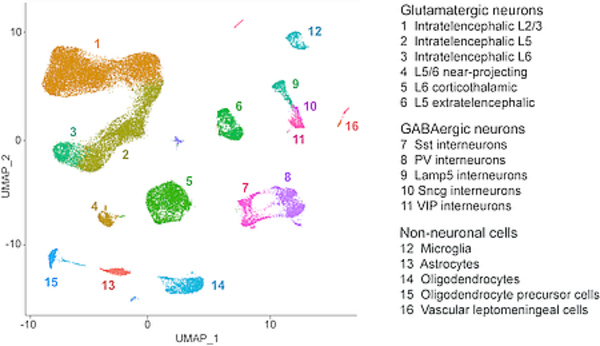
Figure 1.
Two-dimensional projection of nuclei (color-coded by cell type) from the mouse primary motor cortex, analyzed by single nucleus RNA sequencing.
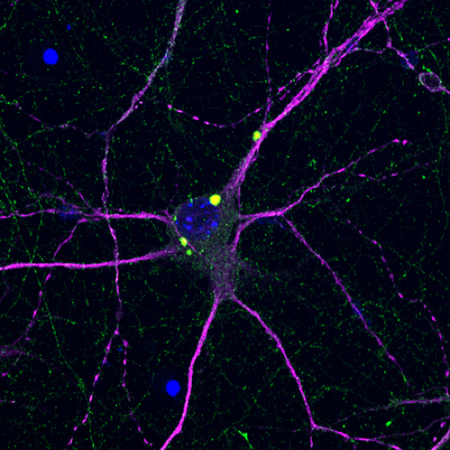
Figure 2.
Murine primary cortical neuron transfected with mutant Huntingtin (mHTT, green). Neuronal marker MAP2 (magenta) shows cellular morphology, nucleus is labelled in blue. Note aggregates of mHTT in the cell body and in neurites.
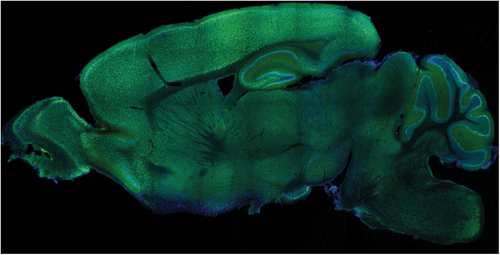
Figure 3.
Sagittal section of a mouse brain neonatally injected with a viral construct to express a YFP-tagged candidate gene (green) in the nervous system. Nuclei are labeled in blue. Note the broad distribution of the ectopic protein in the brain.